1. Introduction
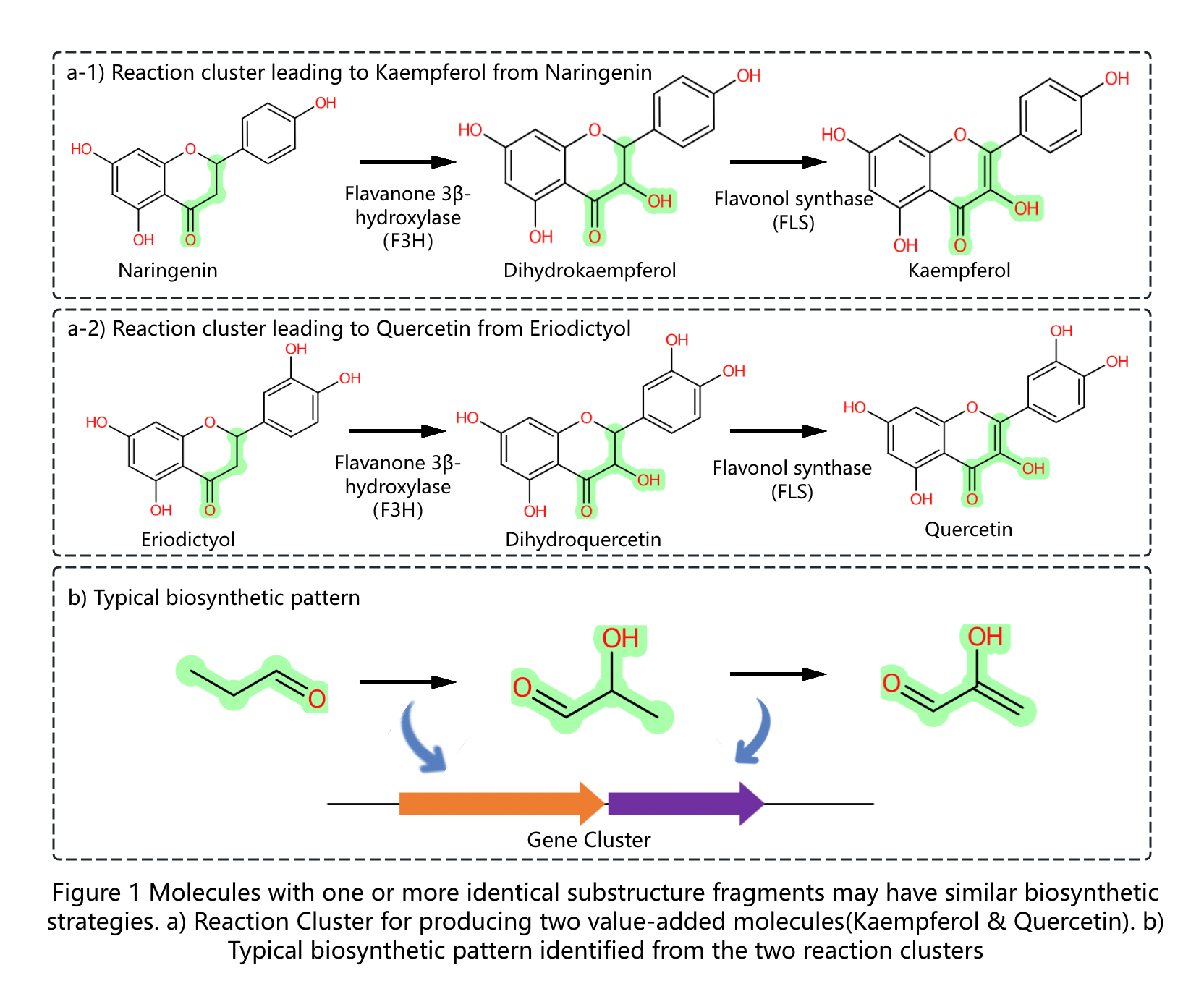
Biosynthetic pathways for producing target molecules can be regarded as series of sequential reactions that can also be digitalized as typical biosynthetic patterns (reaction rule clusters) for producing analogs. Conventional methods for pathway design in silico consider only reaction rules with a single step, which neglect the more efficient synthetic strategies crossing multiple steps. The structure of a molecule is topological and can be divided into multiple substructures, different molecules with one or more identical substructure fragments may have similar biosynthetic strategies(Figure 1). Here, based on the concept of gene clusters, we constructed a user-friendly platform (RxnCluster) by digitalizing the typical biosynthetic patterns for the first time. RxnCluster contains 14,378 biosynthetic patterns (reaction rule clusters) covering 37,317 reaction combinations (reaction clusters) whose numbers of steps vary from 1 to 4. According to the results, this platform can identify the reaction clusters in various numbers of steps, which are consistent with the experimental results obtained in wet laboratories. In addition, it can identify other novel reaction clusters that have not yet been reported, which will pave the way toward pathway mining for molecule biosynthesis via different strategies.The workflow of this platform is shown in Figure 2.
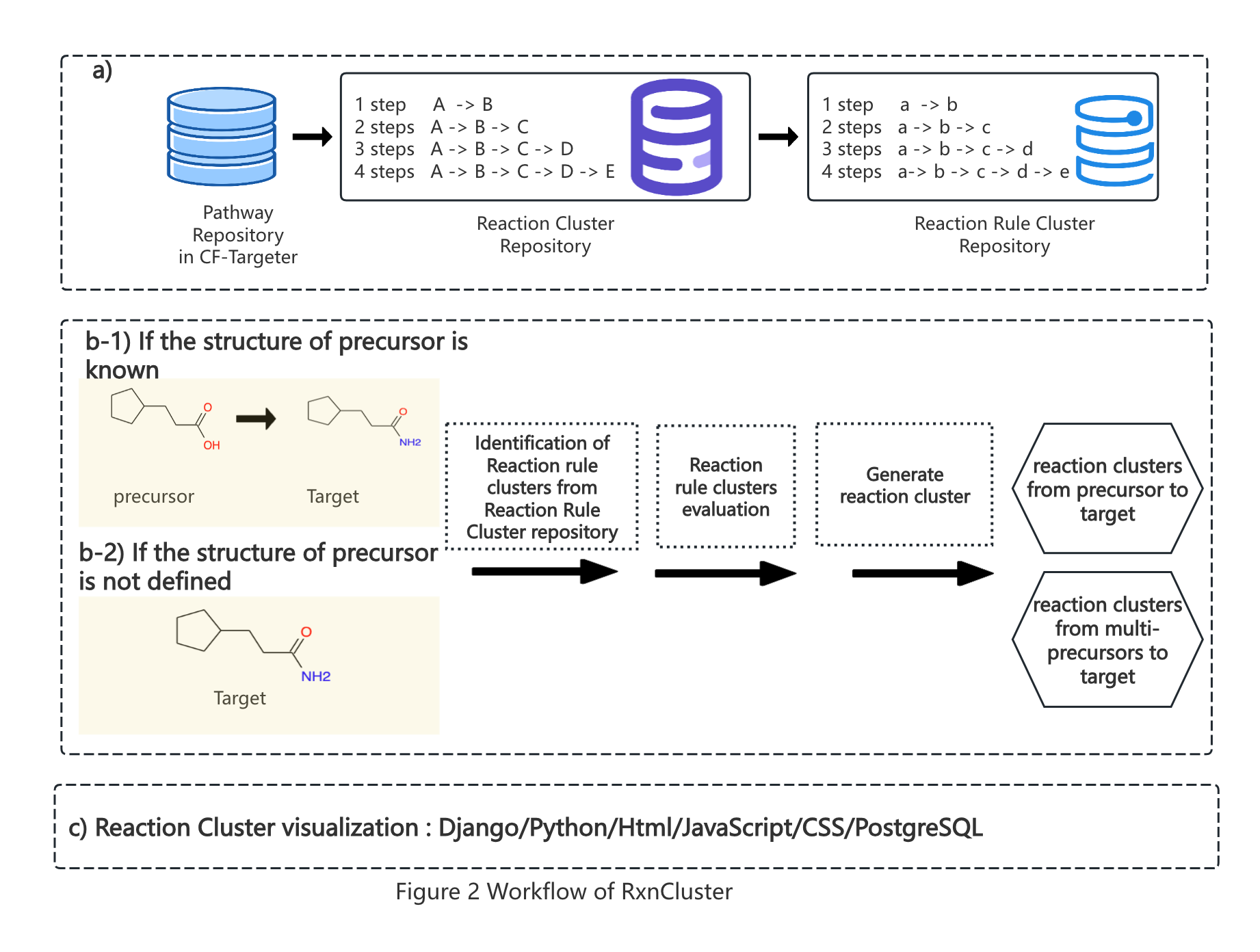
2. RxnCluster Implementation
RxnCluster is developed on the basis of the Linux, NGINX, PostgreSQL, Python strategy.The server runs under NGINX on a Linux machine running Ubuntu Server. The algorithm and backend program were written in Python by using the Django framework in combination with PostgreSQL to manage the data. Hypertext Markup Language (HTML), Cascading Style Sheets (CSS), and JavaScript were used to implement RxnCluster’s front-end data presentation and interactions.3. Draw Molecule
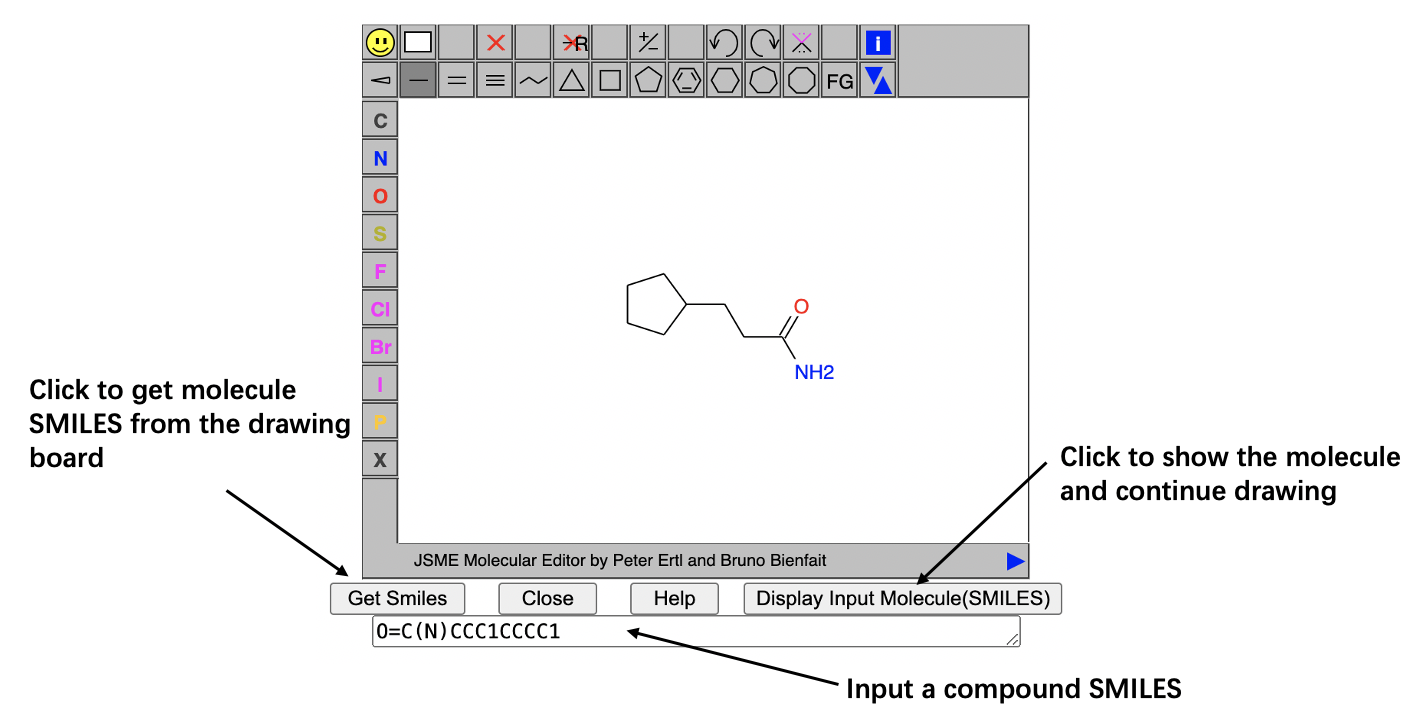
User can draw a molecule from the blank drawing board or start drawing molecule from a structure that has already in the text box.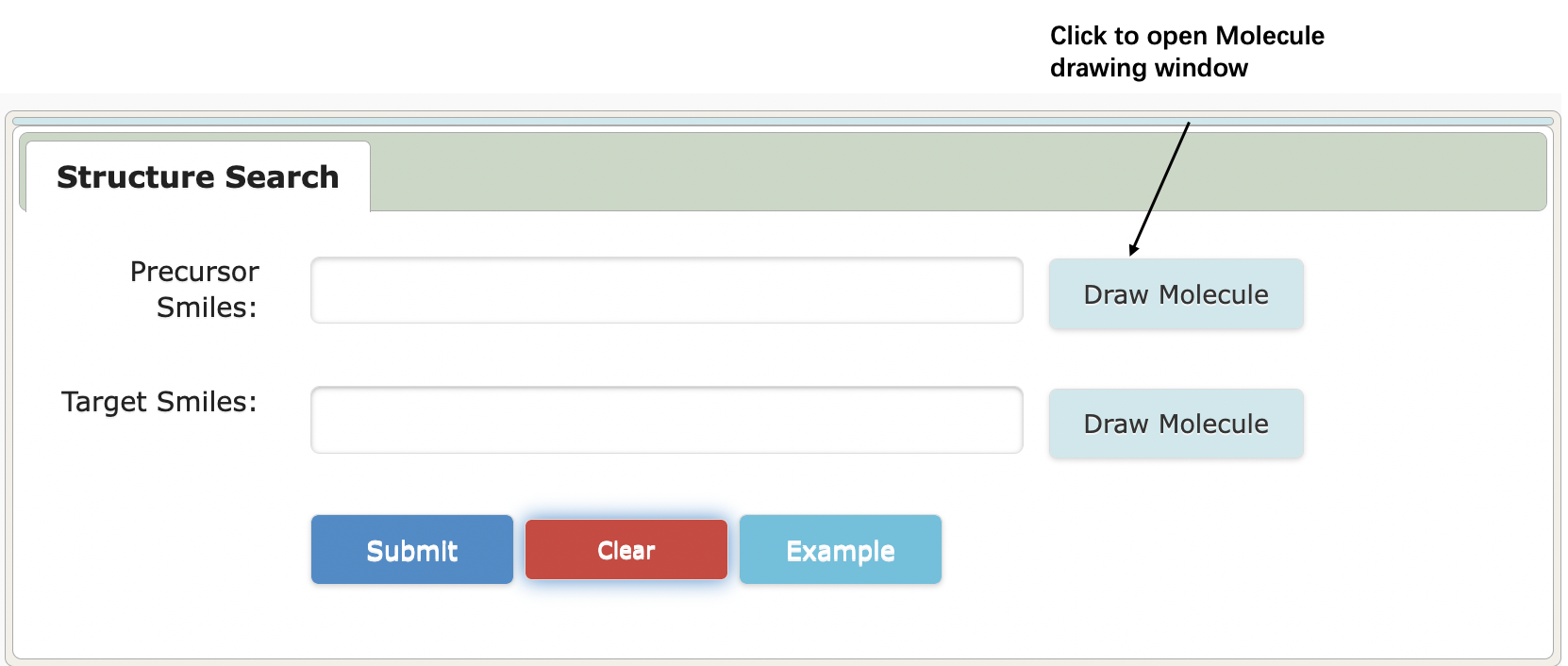
4. If both precursor and target are known
4.1 Input/Get Molecule SMILES
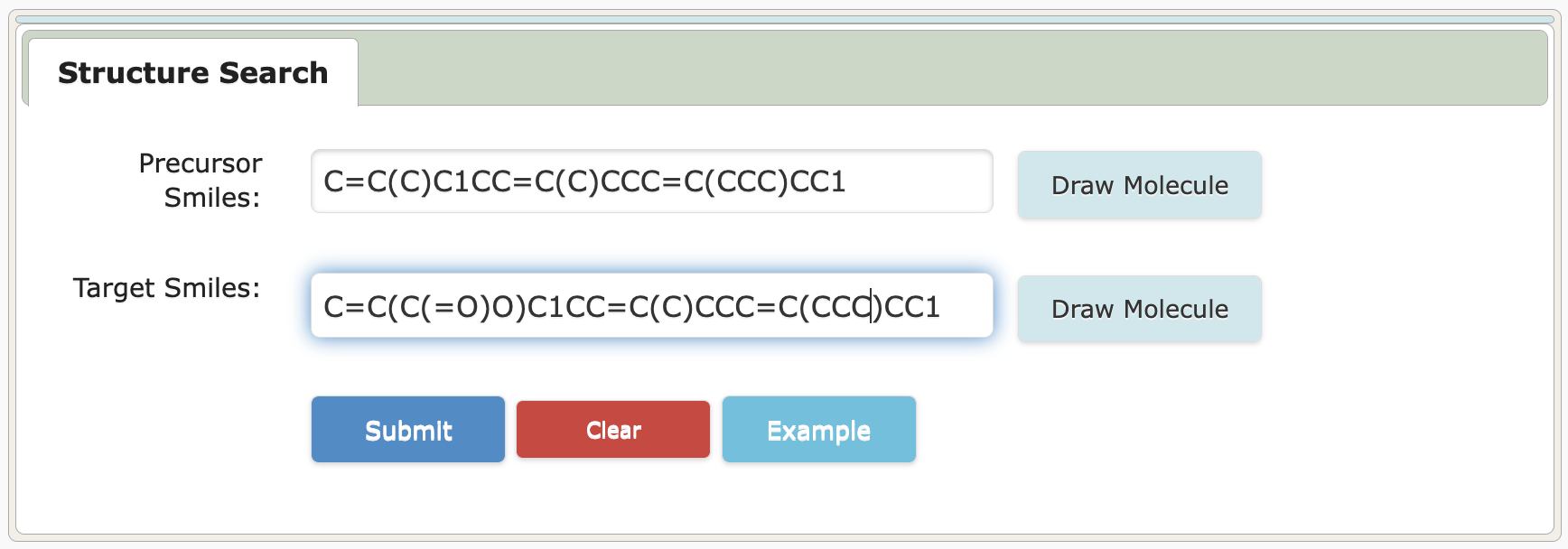
4.2 Get Rule/Reaction Cluster
Get biosynthetic patterns and predict reaction clusters between precursor and target molecule under mult-steps which step length varying from 1 to 4.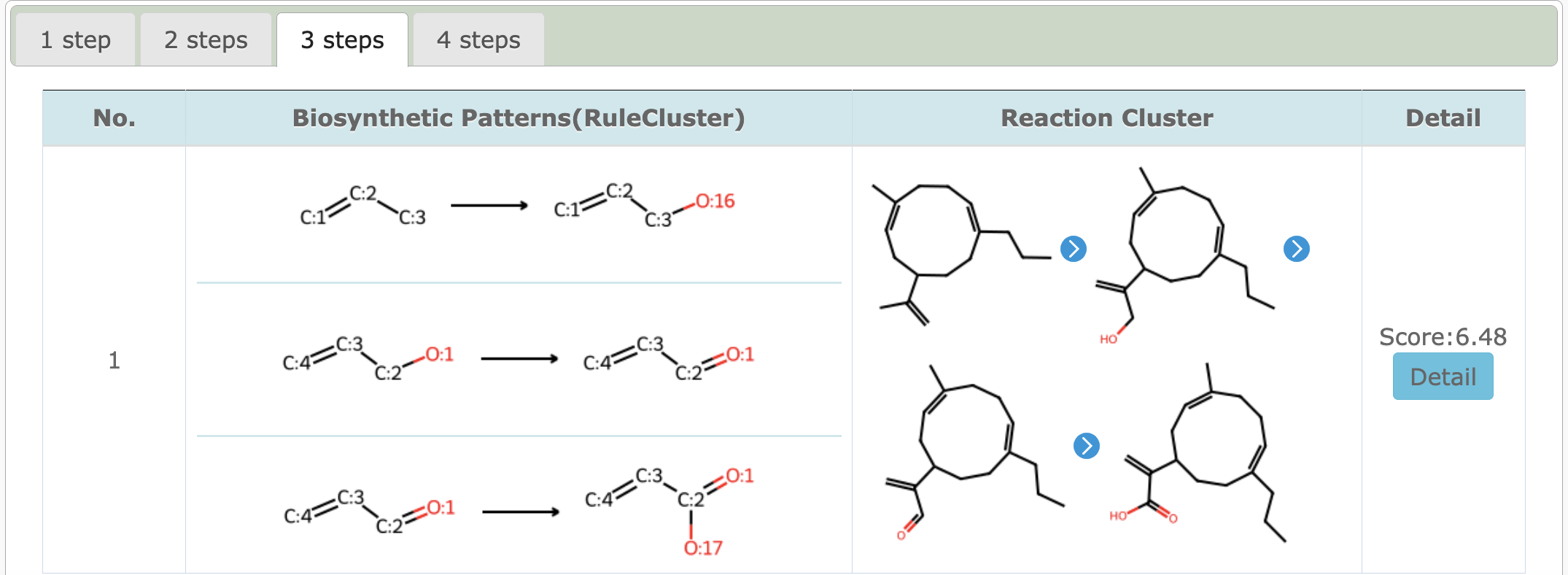
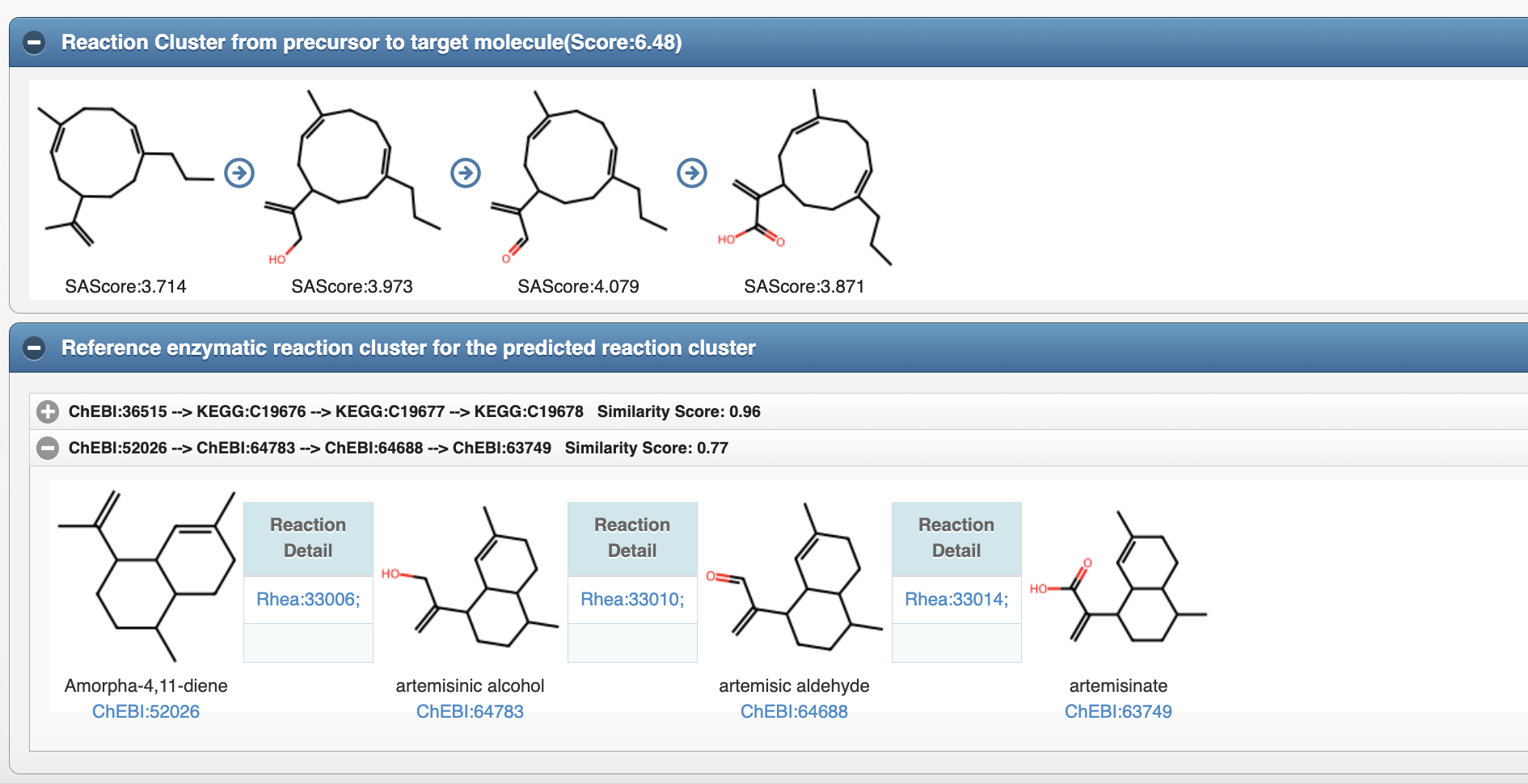
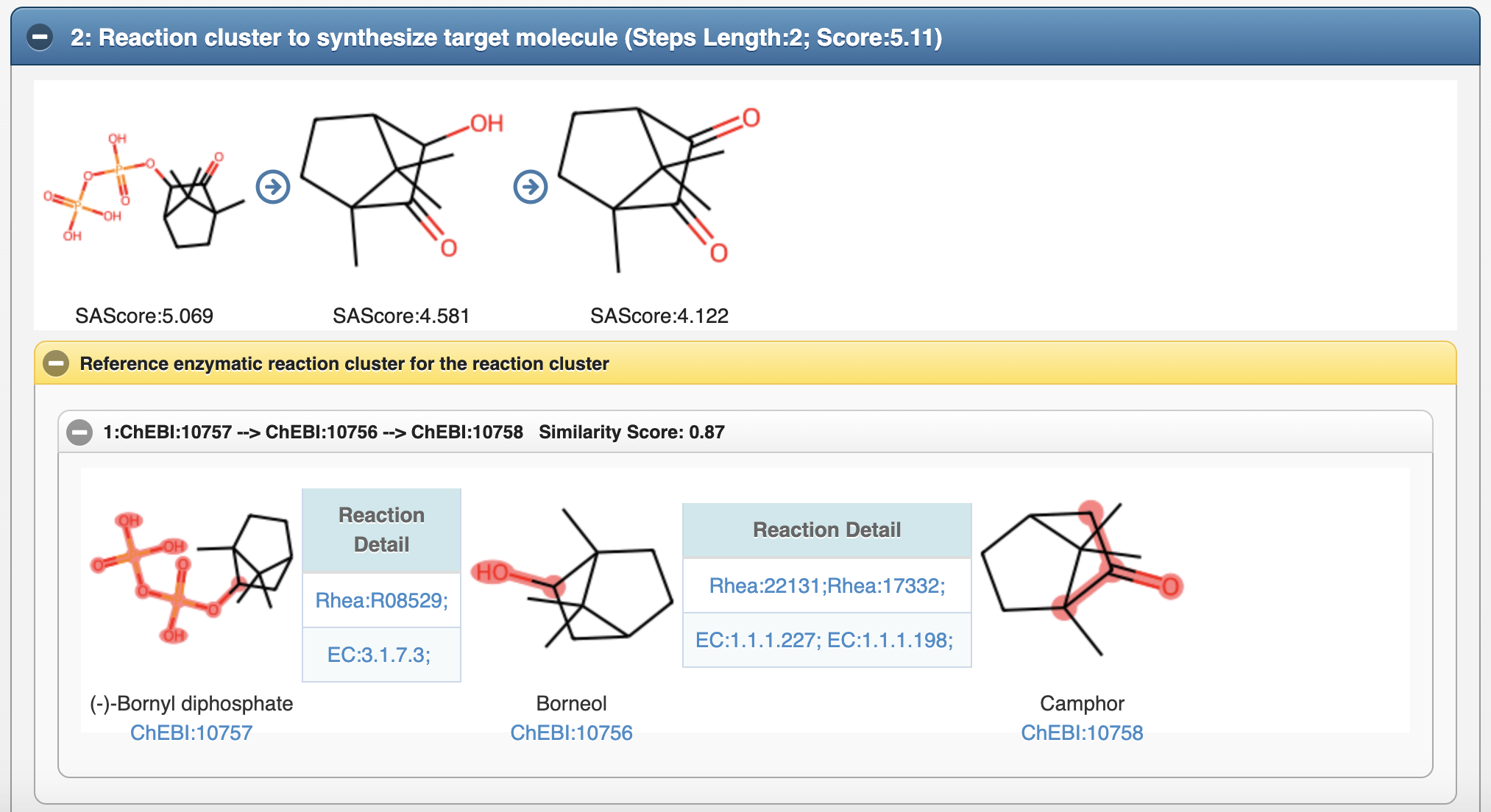
4.3 Reference enzymatic reaction cluster for predicted reaction cluster
5. If only the target molecule is known
5.1 Input/Get target molecule SMILES
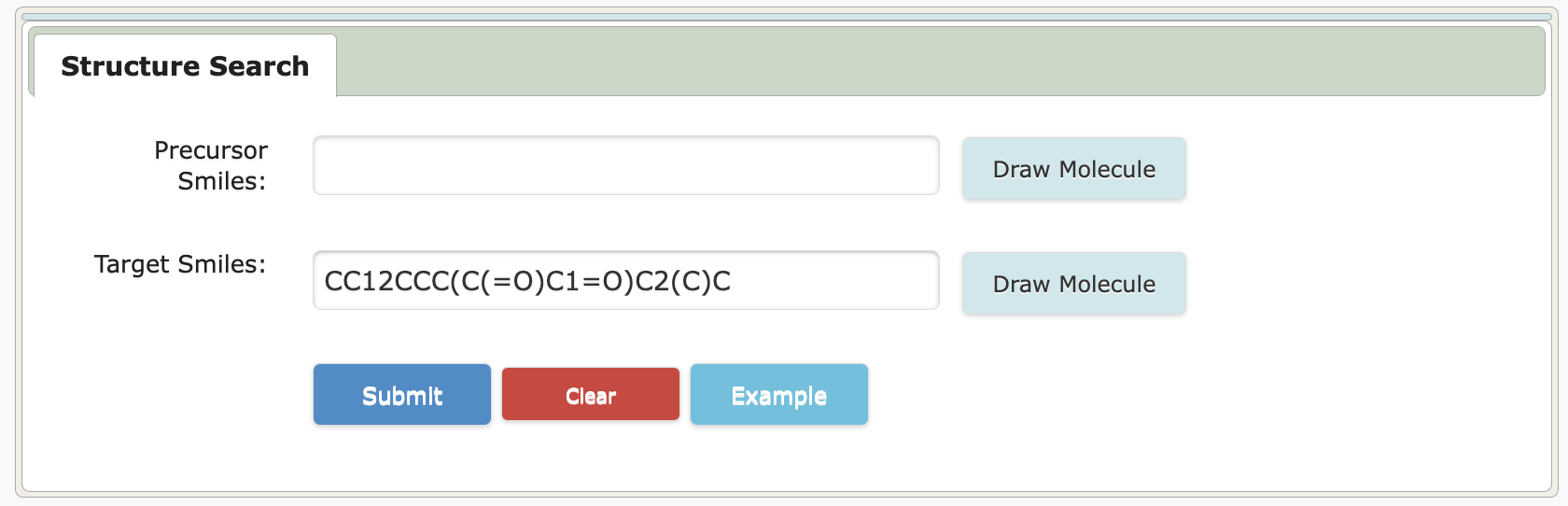
5.2 Search similar compounds
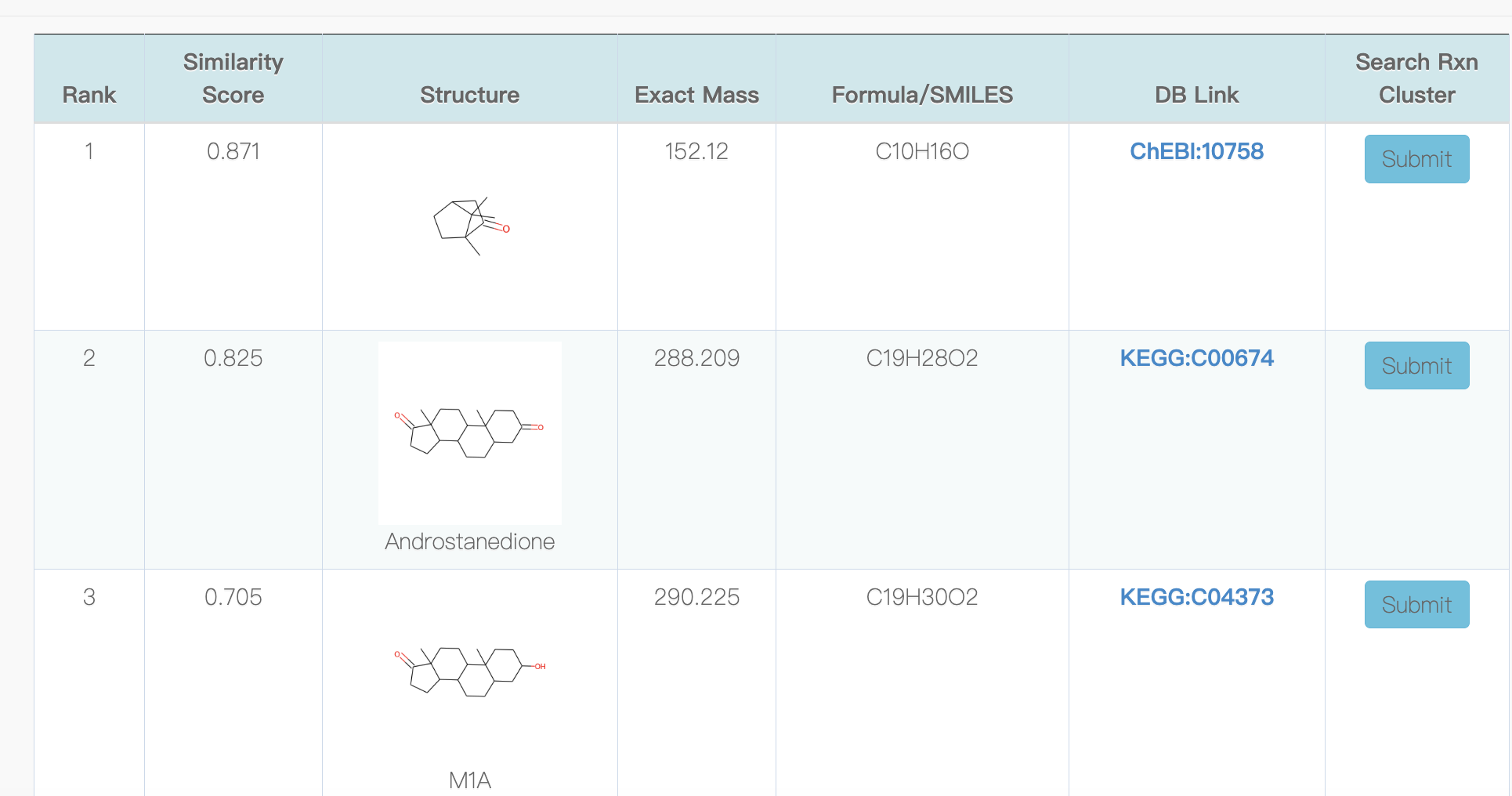
5.3 Reference enzymatic reaction cluster for the predicted reaction cluster
6. Meaning of some terminologies
(1).SMILESThe simplified molecular-input line-entry system (SMILES) is a specification in the form of a line notation for describing the structure of chemical species using short ASCII strings. You can also enter the molecule into RxnCluster via the JSME molecule editor.
(2).Similarity Score
Molecular similarity Score is range from 0 to 1, which reflect the similarity between target molecule and molecules that have biosynthetic patnways in CF-Targeter.After user select one from the list of target analogues, reaction clusters toward target molecule will be predicted on the basis of biosynthetic patterns existed in pathways. The similarity score is calculated by RDkit.
(3).Typical Biosynthetic Patterns
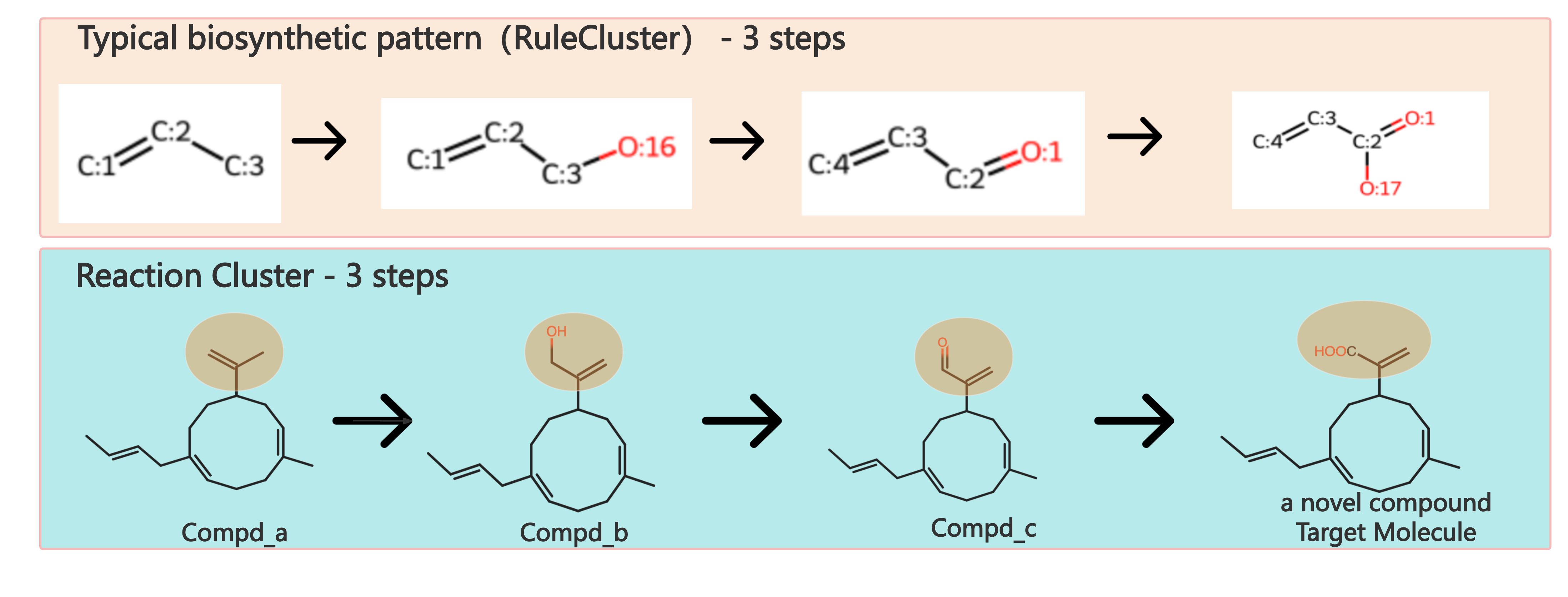
Biosynthetic Pattern(Reaction Rule Cluster) is composed of sequential chemical-structure changes in reaction center.The concept of reaction rule has been used in our previous work(Nucleic Acid Research,2020), which represented by SMARTS.Biosynthetic Patterns are extracted from biosynthetic pathways in our previous work (CF-Targater), and they can be regarded as typical biosynthetic strategies for producing molecules which have same substructures with previous studied molecules.For example, the reaction cluster(a-1) leading to artemisic acid from Amorpha-4,11-diene as well as the reaction cluster(a-2) leading to Gemacrene A acid from Germacrene A can be summarized as synthetic patterns(Figure b) with 3 steps.

(3).Reaction cluster
Based on the typical biosynthetic patterns extracted from biosynthetic pathways, reaction cluster from precursor to target molecule could be predicted.
7. Frequently asked questions
Can I use the RxnCluster data in my research?RxnCluster is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, not requires permission of the authors.
What if I find some error in the database?
Please send an error report to 23113176@whpu.edu.cn and we will verify it as soon as possible. It would be very helpful if you could provide the correct information.
Further questions?
If you have any other questions, please feel free to contact us by email 23113176@whpu.edu.cn